The TOPO cloning technology (compared
with PIPE and standard cloning in ProteoCool n° 1 is a simply and fast cloning approach able to
accommodate a wide range of PCR insert sizes.
TOPO technology enables inserts with
compatible ends to be readily joined to the vector in 5 minutes, without the
need for additional ligation steps.
In my experience in some cases, incorrect Gene insertion (reverse) may happen in a certain % (in my expereince 40-50%) of the E.coli clones obtained from TOPO reaction:
This problem could be generally overcame just by screening and sequencing a large number of colonies (at least 4-6 for each clone)
However is important to pay attention to it during the PCR colony screening and the following sequence check:
For example, PCR colony screening (see ProteoCool n° 4 for more detail about it) performed with primers those anneal in the vector backborne will be not able to differenziate clones with reverse insertion from the good ones.
Since the regions of the TOPO vectors located around (just before and after) the GOI insertion sites are quite similar (especially in the case that the GOI is cloned in a plasmid that do not codify for any C- or N- terminal tag) is possible that the reverse insertion cpuld be not revealed by a not expert users.
Example of cloning of a C-terminal His-tagger GOI in pcdna 3.4 TOPO for recombinant protein expression in mammalian cells (eg Expi293 or ExpiCHO)
In this case primers in addition to the GOI annealing sequence have to contain a flag carryng:
- KOZAC sequence before ATG start codon (Forward primer);
- codons codyfing for the His tag followed by 1 or 2 Stop codons;
The final expected sequence (in the GOI is inserted in the correct direction) will be:
TGACCTCCATAGAAGACACCGGGACCGATCCAGCCTCCGGACTCTAGAGGATCGAACCCTTgccaccATG-GOI-HIStag-STOP -STOP -AAGGGTTCGATCCCTACCGGTTAGTAATGAGTTTGATATCTCGACAATCAACCTCTGG
In yellow --> The region of pcdna3.4 before the insertion,
In Green --> The region of pcdna 3.4 after the insertion;
and the respective Aminoacid traslation will be:
DLHRRHRDRSSLRTLEDRTLATM-GOI-HisTag**KGSIPTG***V*YLDNQPL
where * indicates STOP CODONS
On the contrary in case that the GOI is inserted in the opposite direction the final sequence will be:
ATTTTGTAATCCAGAGGTTGATTGTCGAGATATCAAACTCATTACTAACCGGTAGGGATCGAACCCTTgccaccATG-GOI-HIStag-STOP-STOP-GGGAGGGGGAAAGCGAAAGTCCCGGAAAGGAGCTGACAGGTGGTGGCAATGCCC
and the respective Aminoacid traslation will be:
FVIQRLIVEISNSLLTGRDRTLATM-GOI-HISTag**KGSIL*SPEAGSVPVSSMEVKTAWM
Performing an allingment you can easly see how the insert flanking regions are really similar
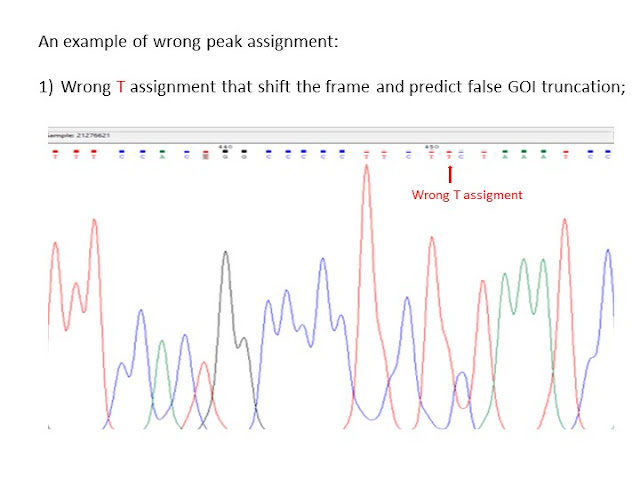
therefore during the sequence check, the sequence analisys cannot be restricted to the few bases close to the insert but need to be extended at least to 10-20bp before and 15-20bp after the insert.
The risk of this kind of mistake is higher in vectors as pcdna3.4 those do not codify for any additional N- or C-terminal tag. In a vector those codify for a TAG, in case of reverse PCR insertion, you will found the AA sequence of the TAG traslated in the opposite direction respect than the sequence of the insert.
In case you have some clone produced using TOPO cloning and do not show any expression,, i suggest to you, before re-design the cloning strategy, to perform a double sequence check to be sure that your gene was inserted in the correct direction.