In the last 20 years several different software packages able to support the scientist in their molecular cloning activities (e.g primer desing, sequence assembly, transaltiion, vector desing desing to manipulate DNA and protein sequences as CloneManager, SnapGene, VectorNTI (discontinued by Thermo from 2019)
Those software are fully integrated set of tools for e cloning simulation, graphic map drawing, primer design and analysis. They are charaterized by a huge number of functions and they require the payment of a licence.
Are those advanced softwares striclty necessary for a molecular biologist that would like just to check the result of a gene cloning into a mammalian or bacterial expression vector ?
In my opinion NO!
Those softwares could be certanilly useful (for example to map the primer annealing region, to desing the vector map) but not essential expecially if you have to produce a limited number of clones in parallel.
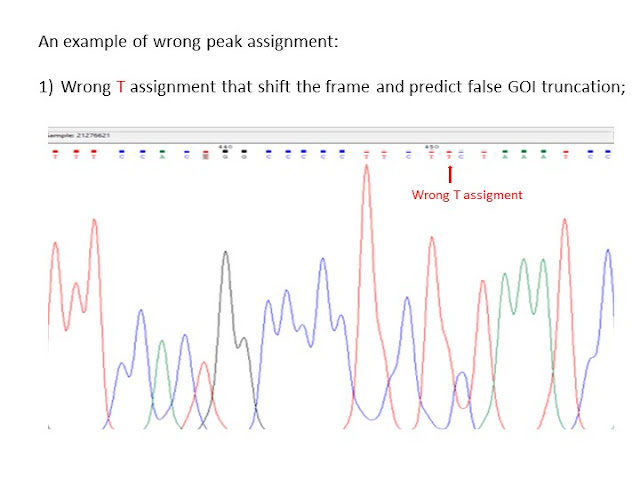
For example, in this post i would like to present youm Chromas, which is a free simple, easy-to-use sequence viewer and editor (able to open chromatogram files(.ab1) produced from automated Sanger sequencers) that could be used to check your sequencing results.







No comments:
Post a Comment