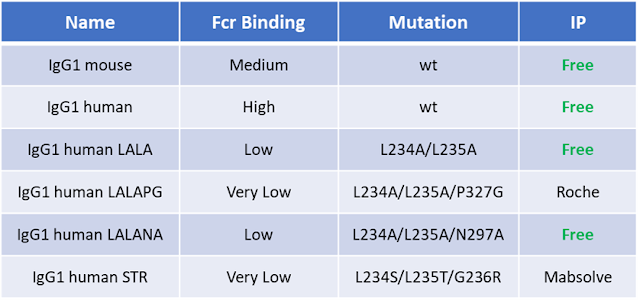
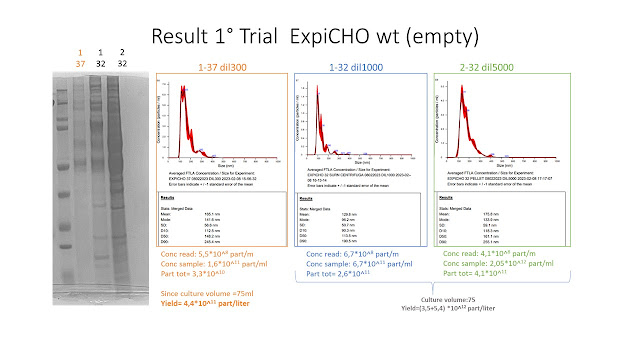
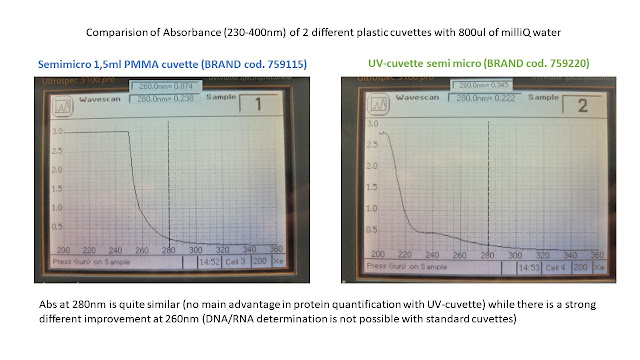
Antibiotic resistance genes (e.g Ampicillin, Kanamycin) are the most commonly used markers for plamisd selection in DNA production and recombinant protein expression processes.
Adding an antibiotic resistance gene to the plasmid solves 2 problems at once: It allows a scientist to easily select the plasmid-containing bacteria when the cells are grown on selective media and at the same time provides those bacteria with a pressure to keep your plasmid.
For the reason the most of commercial vectors suitable for recombinant protein expression in E.coli as pET, pQE, pMal, pCold, pGEx carry Ampicillin, Kanamicin selection marker. .
Even if this strategy work very well in R&D setting, on the contrary it has several drawbacks for biological manufacturing.
The spreading of antibiotics in the environment and consequent emergence of multi-resistant pathogenic bacterial strains has become a general promise to even further increasome categories of biological products such as DNA vaccines where potential issue of allergic reesponses to some classes of antibiotics is evoked and the necessity to document the trace amount of antibiotics.
Apart from the use of the antibiotic itself, there is another emerging limitation in the use the antibiotic resistance gene used as a selection marker due to the potential risk of horizontal transfer of antibiotic resistance gene to environmental microbes
Behind the regulatory issues, the use of antibiotic resistance gene marker imposes a significant metabolic burden on the cells and may. also impact the process yield.
At least but not last,antibiotics themselves are expemsive and therefore often omitted in fermentations, leading to plasmid loss and a corresponding loss in product yield.
In this context novel strategies to efficiently replace antibiotic-based selection are required.
To date, several systems have been developed based on different principles, each presenting advantages and drawbacks.
The most common way to achieve selection in the absence of antibiotics is the complementation of an essential gene making use of an expression vector in a strain with a defect or inhibited expression of the same essential gene.
Several examples are reported in literature where the plasmid selection is achieved through the complementation of amino acid auxotrophy (e,g Proline; Glycine) and more recently of QAPRTase, an enzyme implied in de novo nicotinamide adenine dinucleotide biosynthesis.
Many different E.coli auxothopic strains were already generated but to ensure the proper selective pressure, the auxotrophy complementation systems reported so far require the use of chemically defined media since the standard complex media contain variable amounts of aminoacids as well as other catabolites that covercome the need of biosynthetic pathway and as a consequence the loss of selective pressure for the complementation plasmid maintenance.
About 10 years ago I was involved in a project aimed to developed a novel antibiotic free plasmid selection approach for protein selection in E.coli.
Some experiences gained in previous work activities such as:
1) The positive effect that the supplementation δ-ALA (δ-aminolevulinic acid) has on recombinant Human cyt c expression in E.coli (CERM)
2) The use of 5-aminolevulinic acid (ALA) as a prodrug to stimulate intracellular Heme biosynthesis to produce the natural photosensitizer (PS) Protoporphyrin IX (PpIX) in antimicrobial photodinamic therapy (Molteni Therapeutics)
lead me tot think that the complementation of hemA deficient bacteria by a vector carrying a functional hemA gene, as a selection marker, which confers to the transformed bacteria the ability to grow and maintain the vector in any medium that does not support growth of hemA deficient bacteria of a mutation in heme biosynthetic pathway and at the same time the complementation of empty cells with 5-ALA allow to easly propagate and prepare the empty competent cells.
After several months of work we produced an interesting data package supporting our hipotesis and a patent application was filled and submitted (WO2015165841 - AN ANTIBIOTIC-FREE METHOD FOR SELECTWO2015165841 - AN ANTIBIOTIC-FREE METHOD FOR SELECTION OF TRANSFORMED BACTERIA)
Even if the patent was not accepted since the examing authority do not recognize its inventive steps since the HemA was already show to work as selectable marker in Aspergillus Oryzae, i still think that it is work very well in E.coli since we was able to show that the Delta HemA E.coli show negligible growth in both chemical defined and complex media but the growth it readly restored when there are complemented with 5-ala or plasmid carryng (see Fig2a,2b and Fig3a,3b of the WO2015165841 patent application) the HemA gene and that the expression was mantained after a several different passages.
E. coli BL21(DE3) DeltaHemA/pet21-BFP and E. coli BL21(DE3)/pet21-BFP were cultivated without antibiotic selection for many generations by diluting a 12h culture 1:100 in fresh medium for several times. After 1,2,5,10,15 passages the recombinant protein expression was analyzed (1G ≈ 7-8 duplications) and as reported in the Fig5 of the WO2015165841 patent apprication, the BFP production in HemA mutant strain complemented with BFPpet24-HemA is stable after 15 cultivation cycles without antibiotic while in the wild type strain transformed with standard BFPpet24, it dramatically decreases after 5 cultivation cycles without selection.
I would like to Thanks to
Maria Giuliani
that performed the most of the experimental work
and provide an essential contribute in experiment desing